Datasets
We endeavor to make the important components of data from key analyses available in addition to code repositories.
Mostly we house these as zenodo.org, but in the future, may take advantage of newly available capabilities at Penn.
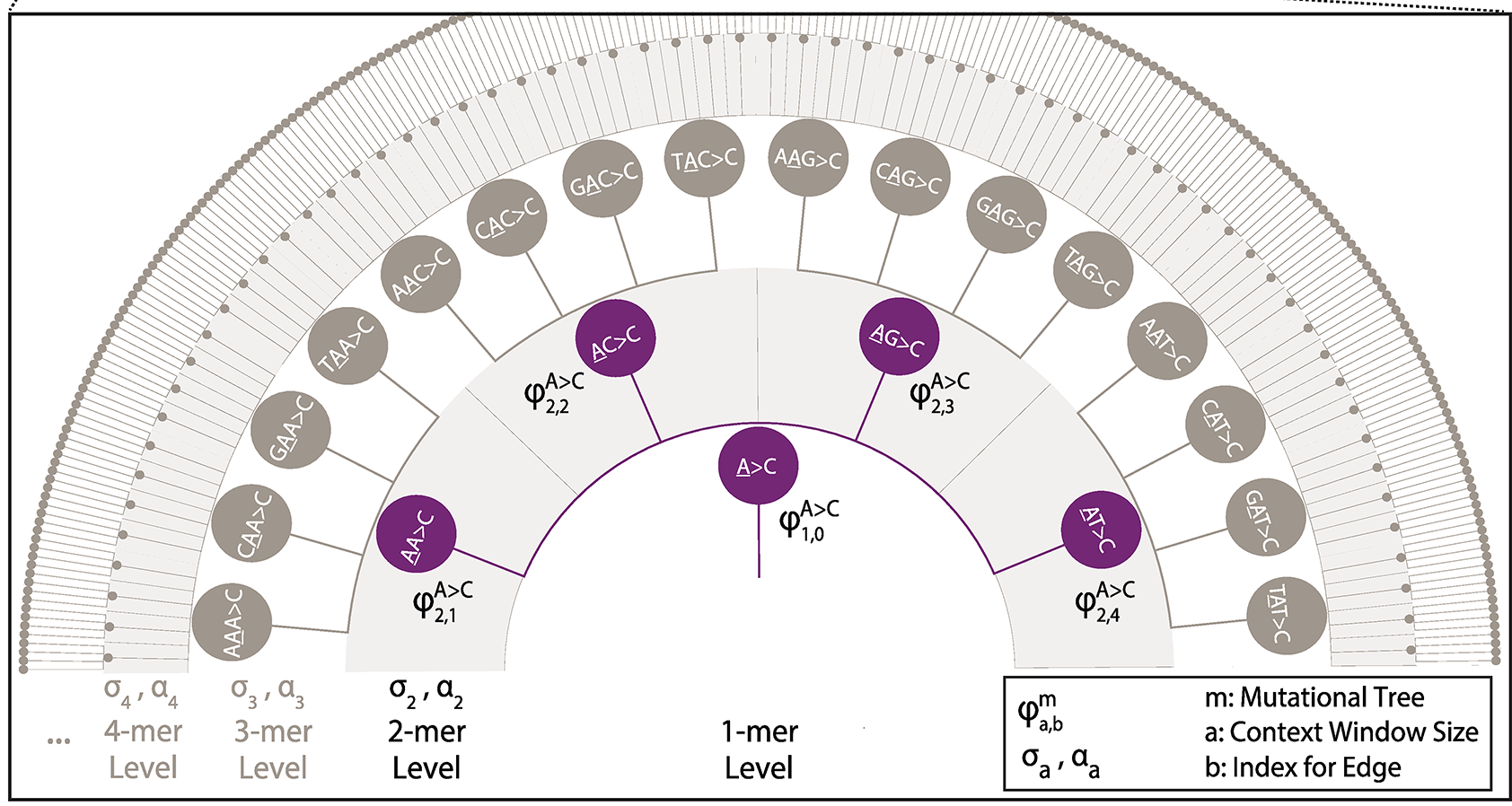
Additional data on outputed models from Bayer as reported in Adams et al (2023).
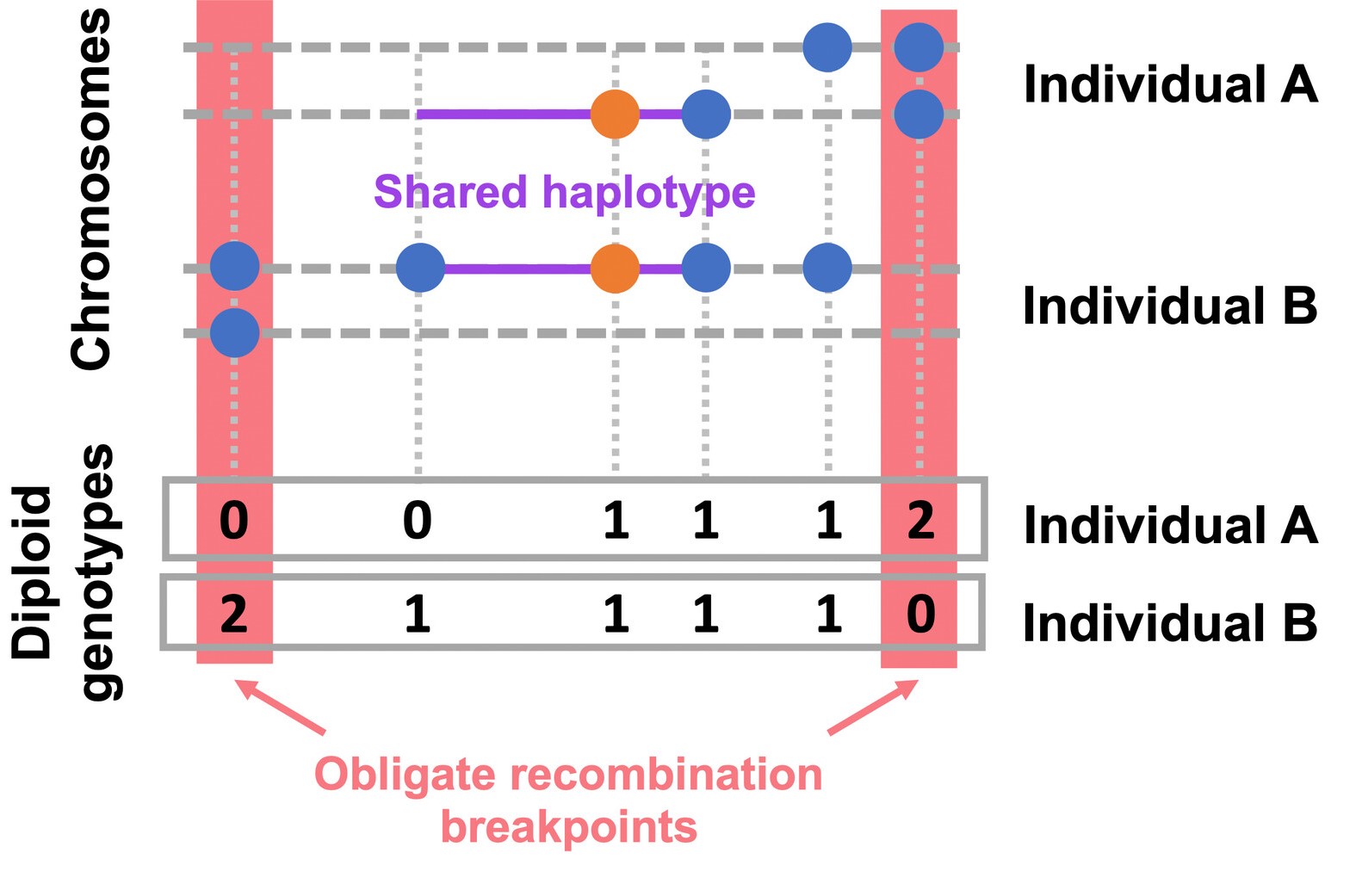
Simulation output and Genome-wide scan for nIBD variants in UK10K data as reported in Johnson et al (2022).
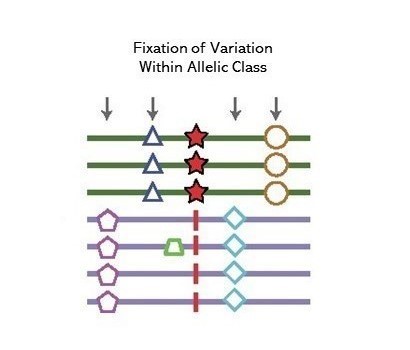
Genome-wide and top 1% scores for 1KG project data output from BetaScan reported in Siewert and Voight (2017).

Genome-wide scan using BetaScan2 in 1KG populations report in Siewert and Voight (2020).
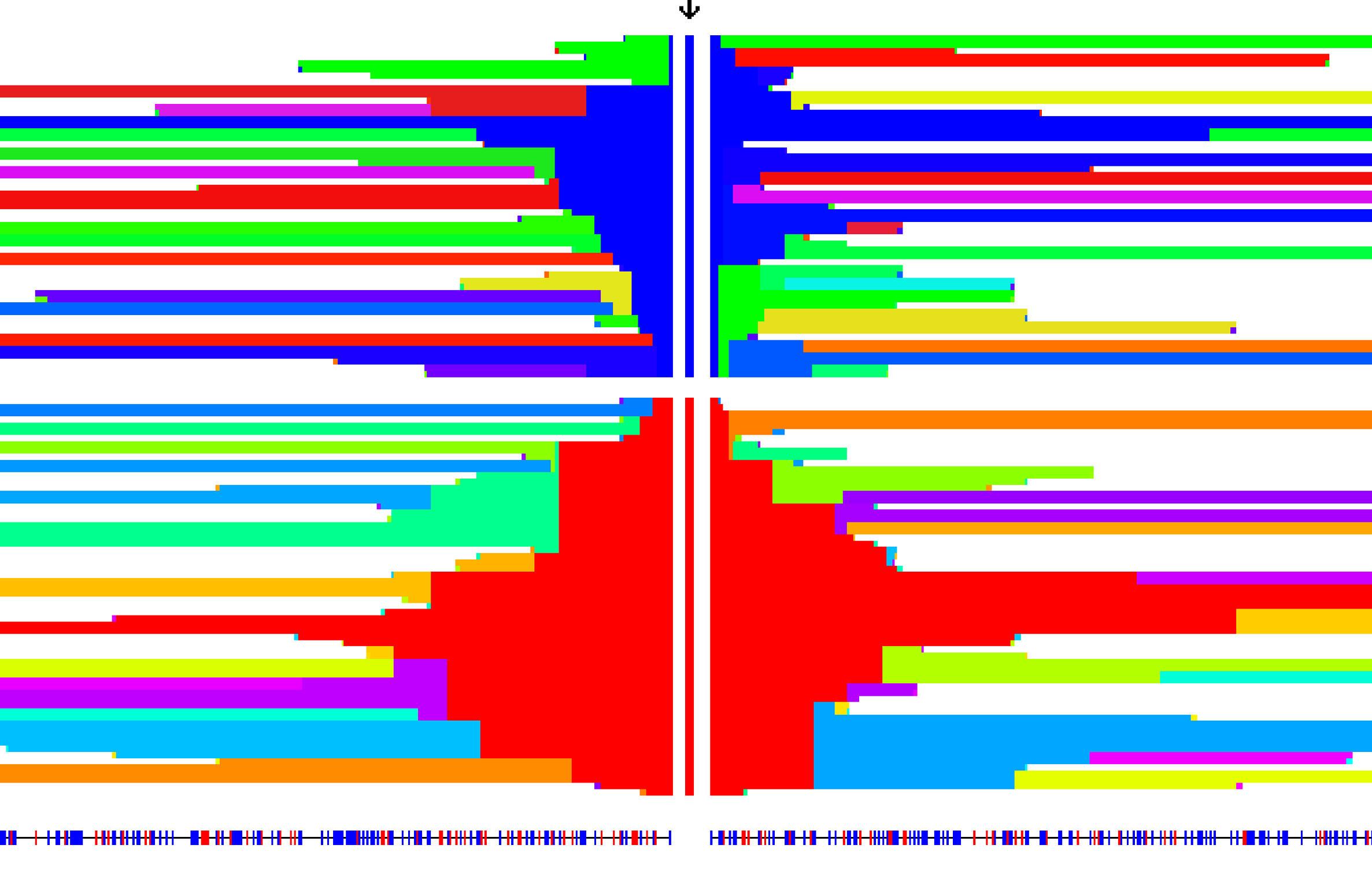
Genome-wide summary stats for modified iHS scan in 1KG as reported in Johnson and Voight (2018).

Summary of Bivariate GWAS scan results reported in Siewert et al (2018).