Research Interests and Scientific Activities

Population Genetics and Evolution
We are an active population genetics lab and projects focus around studies of mutation rate variation, natural selection, and at the interface between population genetics and complex traits. Projects include both methods development and/or data science analysis. We are especially interested in development and applications using Baymer, a tool to infer mutation rate variation based on local nucleotide sequence context that we recently reported. We are also active collaborators with other Population Genetics minded groups at Penn! (Image by Stacie L. Bumgarner, PhD - Red Windmill Studio)

Dissection of Complex Traits
We are interested in the architecture of complex traits, identification of causal genes, and inference of perturbations informed by human genetics predicted to be therapeutically actionable. Our space of primary interest include cardiometabolic traits, including type 2 diabetes, cardiovascular disease, liver disease, and causal traits related to these enpoints. We are also generally interested in pleiotropy at large. We touch all layers, including GWAS, pheWAS, fine-mapping, variant-to-gene mapping, risk prediction via PRS, and collaborations to validate causal genes. (Image credit to Uffelmann et al. 2021, Fig. 3b)
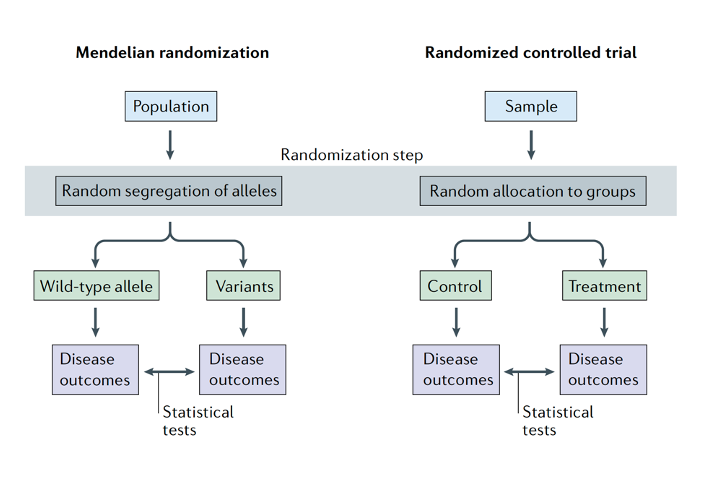
Mendelian Randomization
Causal inference is one of the most challenging - but important - problems in biology and medicine. As a human genetics lab, we are active in the application of Mendelian Randomization as a statistical approach to provide one line of evidence on the causal inference problem. Available projects include Big Data analysis or methods development. (Image credit to Sanderson et al. 2022, Fig. 1)
Consortium Level Activities
My group is active in several consortia, generally focused around human genomics data. The objectives of these interactions are to either assemble data for discovery, collaborate to advance functional validation of causal genes for T2D, access to large-scale biobank data (e.g., MVP), or to develop new knowledge portals to aggregate data for discovery and inference of disease processes.
